Aurora™ 3D Spatial Biology Solution
Ground truth 3D tissue imaging, AI-powered analysis, and a complete workflow for intact tissue volumes.
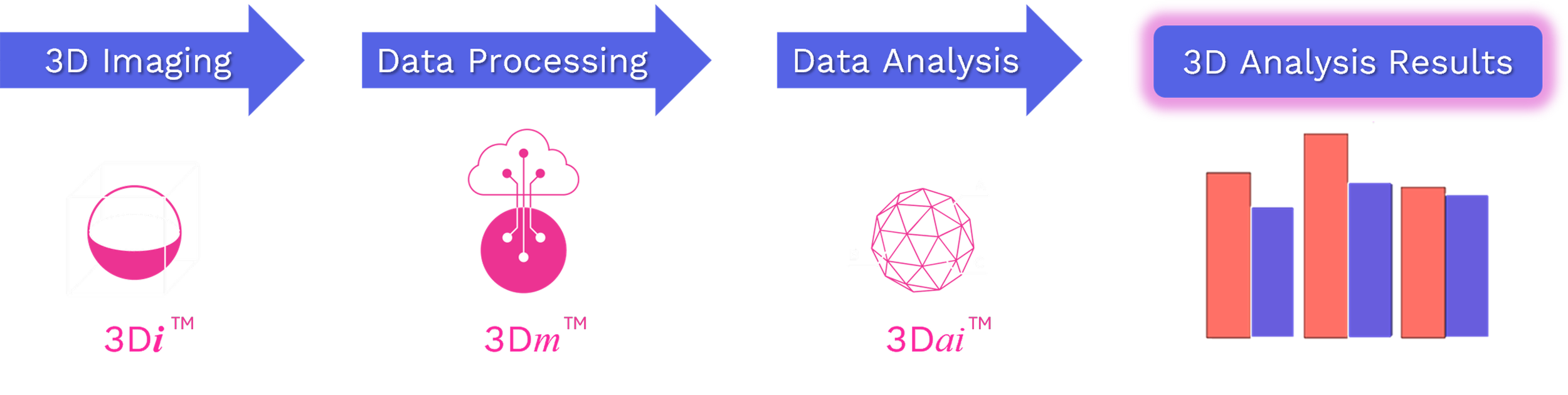
How the Aurora 3D™ Spatial Biology Platform Works
The Aurora 3D™ Spatial Biology Solution is a unified platform that converts intact tissue into quantitative 3D datasets. It includes three integrated modules.
3Di™ is the proprietary light-sheet microscope for high-resolution 3D tissue imaging.
3Dm™ handles automated data management and processing for large volumes.
3Dai™ provides AI-driven image analysis for full tissue quantification.
Aurora 3D™ Platform can be accessed in multiple ways. The 3Di™ light-sheet microscope is available for purchase as a standalone instrument or can be combined with 3Dm™ and 3Dai™ to create an in-house workflow. The whole platform is also available through Alpenglow’s 3D histology imaging services, providing complete imaging, processing, and analysis.
3D Tissue Imaging: 3Di™ HOTLS
High-resolution whole tissue imaging with Hybrid Open-Top Light-Sheet Microscopy
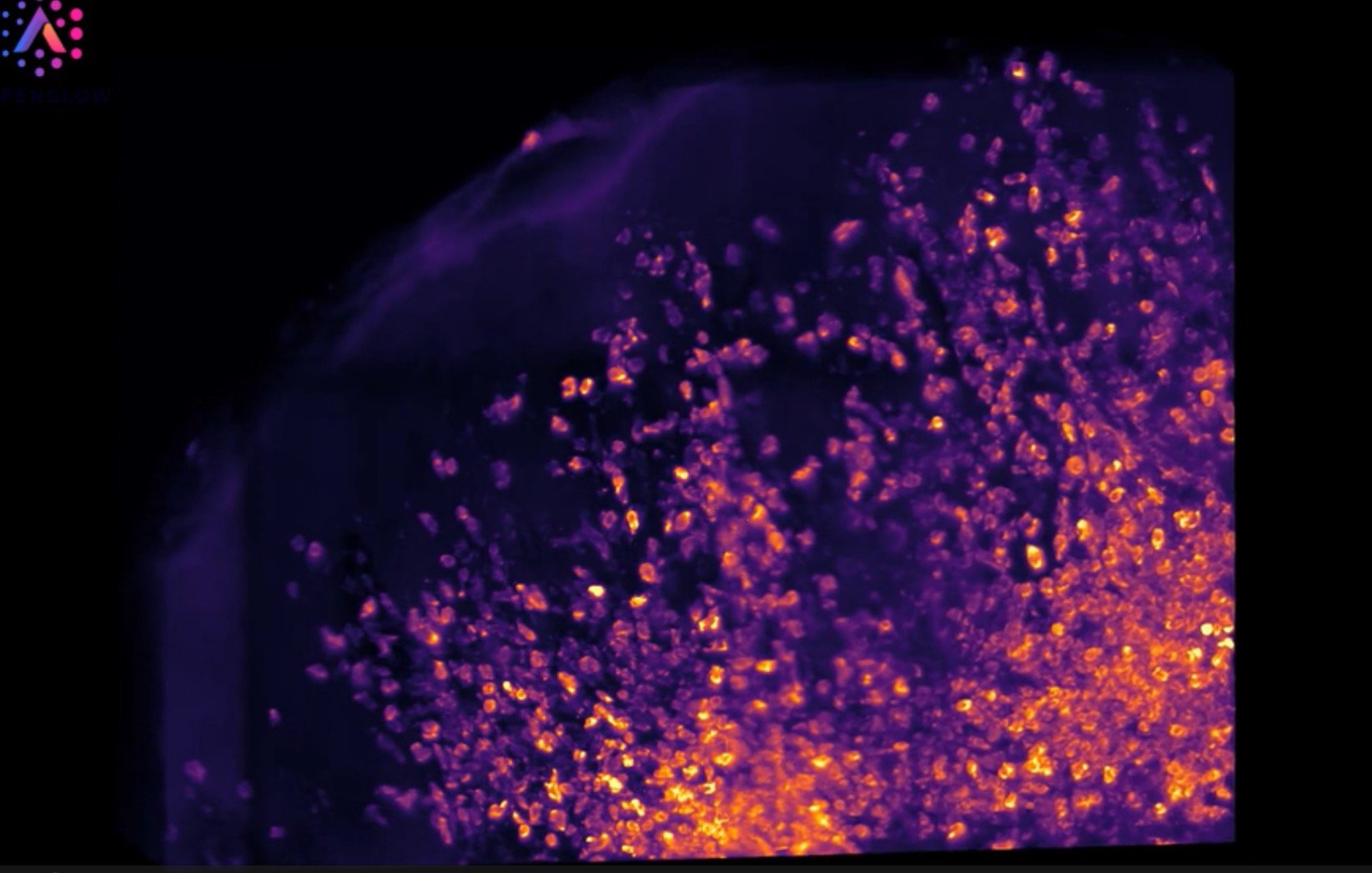
The Aurora 3D™ Platform includes the 3Di™ Hybrid Open-Top Light-Sheet (HOTLS) Fluorescent Microscope. It captures intact tissue blocks using structured illumination and multi-scale imaging, generating clean, high-resolution, submicrometer-detail volumetric datasets. The system supports a 1 cm working distance and scan dimensions of 12.5cm x 7cm as described in the Nature Methods publication. It delivers non-destructive fluorescent imaging across skin biopsies, tumors, lymph nodes, organs, and preclinical models.
The 3Di™ workflow uses scout to zoom imaging. Scout mode surveys large fields quickly to locate regions of interest, then zoom mode acquires high-resolution data with finer sampling for detailed analysis. The open-top architecture simplifies sample handling, improves mounting stability, and accommodates diverse tissue geometries.
Data Management: 3Dm™
Automated registration, alignment, and flat field correction for large 3D data sets
3Dm™ organizes and processes large 3D tissue volumes generated by the 3Di™ system. It manages stitching, illumination correction, multi-channel integration, and volume optimization. The pipeline is designed for fluorescent imaging data and supports multi-scale datasets that span scout and zoom acquisitions. This comprehensive tool automatically aligns images post-imaging, surpassing the speed and efficiency of open-source options, and is validated for data sizes exceeding 4 terabytes.
Image Analysis: 3Dai™
AI powered quantification for intact tissues in true 3D
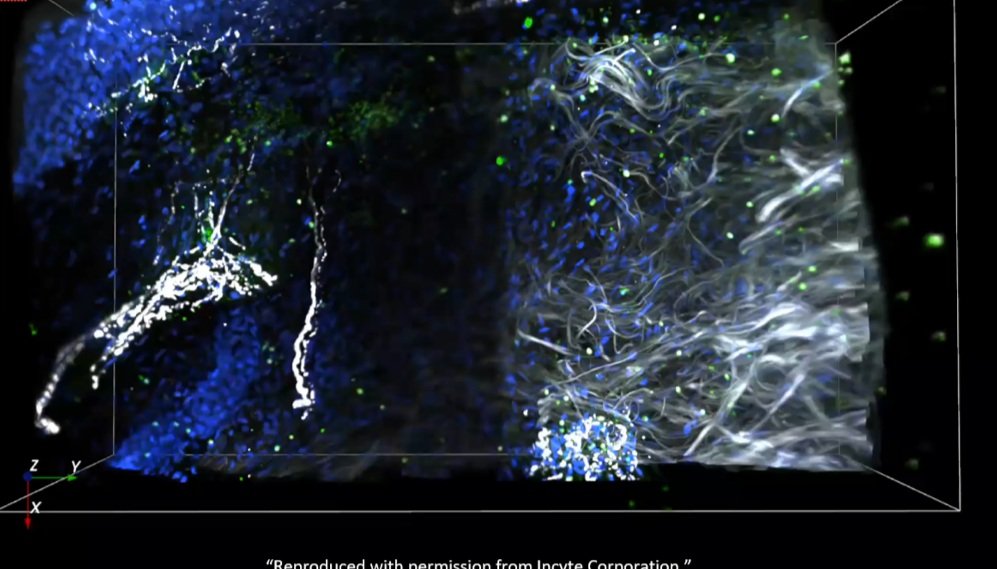
3Dai™ image analysis tool extracts biological meaning from whole tissue datasets using AI-driven analysis. It identifies cells and structures across the entire volume, measures spatial relationships, and provides 3D metrics that are not accessible from traditional 2D sections.
Analysis can include cell segmentation, immune profiling, nerve and vessel quantification, volumetric measurements, region-level stratification, and multi-channel spatial mapping. The platform delivers analysis-ready outputs that integrate with biomarker discovery, translational research, and computational modeling.
Why Aurora 3D™
A platform built for whole tissue imaging and quantitative spatial biology
Aurora 3D™ Platform provides:
Whole tissue imaging for intact samples
High-resolution volumetric datasets
Quantitative spatial profiling
AI-powered 3D analysis
End-to-end workflow from imaging to results
Data quality suitable for discovery, translational programs, and computational pipelines
Aurora 3D™ Platform Applications
Aurora 3D™ Platform supports programs where tissue architecture, cell distribution, and microenvironmental context are essential:
Immunology
Neuroscience
Toxicology
Preclinical research
Clinical research
Spatial biology
Biomarker discovery
Ready to explore the Aurora 3D™ Platform?
Dive into a compelling example of the complete Aurora Workflow, featuring an advanced 3D imaging study of heart architecture.
This video highlights groundbreaking insights into murine cardiac vasculature, offering detailed analyses such as cell counts, vessel dimensions, distance measurements, and branching patterns.